Signal Processing - 101¶
!git clone https://github.com/amirberenji1995/damavand
Cloning into 'damavand'... remote: Enumerating objects: 314, done. remote: Counting objects: 100% (25/25), done. remote: Compressing objects: 100% (18/18), done. remote: Total 314 (delta 8), reused 18 (delta 6), pack-reused 289 (from 1) Receiving objects: 100% (314/314), 9.18 MiB | 24.67 MiB/s, done. Resolving deltas: 100% (152/152), done.
!pip install -r damavand/requirements.txt
Collecting certifi==2024.7.4 (from -r damavand/requirements.txt (line 1))
Downloading certifi-2024.7.4-py3-none-any.whl.metadata (2.2 kB)
Collecting charset-normalizer==3.3.2 (from -r damavand/requirements.txt (line 2))
Downloading charset_normalizer-3.3.2-cp312-cp312-manylinux_2_17_x86_64.manylinux2014_x86_64.whl.metadata (33 kB)
Collecting idna==3.7 (from -r damavand/requirements.txt (line 3))
Downloading idna-3.7-py3-none-any.whl.metadata (9.9 kB)
Collecting numpy==1.26.4 (from -r damavand/requirements.txt (line 4))
Downloading numpy-1.26.4-cp312-cp312-manylinux_2_17_x86_64.manylinux2014_x86_64.whl.metadata (61 kB)
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 61.0/61.0 kB 5.2 MB/s eta 0:00:00
Collecting pandas==2.1.4 (from -r damavand/requirements.txt (line 5))
Downloading pandas-2.1.4-cp312-cp312-manylinux_2_17_x86_64.manylinux2014_x86_64.whl.metadata (18 kB)
Requirement already satisfied: python-dateutil==2.9.0.post0 in /usr/local/lib/python3.12/dist-packages (from -r damavand/requirements.txt (line 6)) (2.9.0.post0)
Collecting pytz==2024.1 (from -r damavand/requirements.txt (line 7))
Downloading pytz-2024.1-py2.py3-none-any.whl.metadata (22 kB)
Collecting rarfile==4.2 (from -r damavand/requirements.txt (line 8))
Downloading rarfile-4.2-py3-none-any.whl.metadata (4.4 kB)
Collecting requests==2.32.3 (from -r damavand/requirements.txt (line 9))
Downloading requests-2.32.3-py3-none-any.whl.metadata (4.6 kB)
Collecting scipy==1.13.1 (from -r damavand/requirements.txt (line 10))
Downloading scipy-1.13.1-cp312-cp312-manylinux_2_17_x86_64.manylinux2014_x86_64.whl.metadata (60 kB)
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 60.6/60.6 kB 5.1 MB/s eta 0:00:00
Collecting six==1.16.0 (from -r damavand/requirements.txt (line 11))
Downloading six-1.16.0-py2.py3-none-any.whl.metadata (1.8 kB)
Collecting tzdata==2024.1 (from -r damavand/requirements.txt (line 12))
Downloading tzdata-2024.1-py2.py3-none-any.whl.metadata (1.4 kB)
Collecting urllib3==2.2.2 (from -r damavand/requirements.txt (line 13))
Downloading urllib3-2.2.2-py3-none-any.whl.metadata (6.4 kB)
Collecting pycatch22==0.4.5 (from -r damavand/requirements.txt (line 14))
Downloading pycatch22-0.4.5.tar.gz (50 kB)
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 50.3/50.3 kB 4.1 MB/s eta 0:00:00
Installing build dependencies ... done
Getting requirements to build wheel ... done
Preparing metadata (pyproject.toml) ... done
Downloading certifi-2024.7.4-py3-none-any.whl (162 kB)
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 163.0/163.0 kB 13.3 MB/s eta 0:00:00
Downloading charset_normalizer-3.3.2-cp312-cp312-manylinux_2_17_x86_64.manylinux2014_x86_64.whl (141 kB)
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 141.9/141.9 kB 13.3 MB/s eta 0:00:00
Downloading idna-3.7-py3-none-any.whl (66 kB)
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 66.8/66.8 kB 4.7 MB/s eta 0:00:00
Downloading numpy-1.26.4-cp312-cp312-manylinux_2_17_x86_64.manylinux2014_x86_64.whl (18.0 MB)
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 18.0/18.0 MB 95.2 MB/s eta 0:00:00
Downloading pandas-2.1.4-cp312-cp312-manylinux_2_17_x86_64.manylinux2014_x86_64.whl (11.7 MB)
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 11.7/11.7 MB 85.2 MB/s eta 0:00:00
Downloading pytz-2024.1-py2.py3-none-any.whl (505 kB)
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 505.5/505.5 kB 33.6 MB/s eta 0:00:00
Downloading rarfile-4.2-py3-none-any.whl (29 kB)
Downloading requests-2.32.3-py3-none-any.whl (64 kB)
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 64.9/64.9 kB 5.4 MB/s eta 0:00:00
Downloading scipy-1.13.1-cp312-cp312-manylinux_2_17_x86_64.manylinux2014_x86_64.whl (38.2 MB)
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 38.2/38.2 MB 13.6 MB/s eta 0:00:00
Downloading six-1.16.0-py2.py3-none-any.whl (11 kB)
Downloading tzdata-2024.1-py2.py3-none-any.whl (345 kB)
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 345.4/345.4 kB 25.8 MB/s eta 0:00:00
Downloading urllib3-2.2.2-py3-none-any.whl (121 kB)
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 121.4/121.4 kB 9.2 MB/s eta 0:00:00
Building wheels for collected packages: pycatch22
Building wheel for pycatch22 (pyproject.toml) ... done
Created wheel for pycatch22: filename=pycatch22-0.4.5-cp312-cp312-linux_x86_64.whl size=113889 sha256=630aa436d9c70aac2eeeae6b26c8d8304814b5a0a4144ba0a80e5d628d39c1f9
Stored in directory: /root/.cache/pip/wheels/89/44/33/6f846a24d9908b045e0e6b7d8a4e66adc4dec8fdf14184c90c
Successfully built pycatch22
Installing collected packages: pytz, pycatch22, urllib3, tzdata, six, rarfile, numpy, idna, charset-normalizer, certifi, scipy, requests, pandas
Attempting uninstall: pytz
Found existing installation: pytz 2025.2
Uninstalling pytz-2025.2:
Successfully uninstalled pytz-2025.2
Attempting uninstall: urllib3
Found existing installation: urllib3 2.5.0
Uninstalling urllib3-2.5.0:
Successfully uninstalled urllib3-2.5.0
Attempting uninstall: tzdata
Found existing installation: tzdata 2025.2
Uninstalling tzdata-2025.2:
Successfully uninstalled tzdata-2025.2
Attempting uninstall: six
Found existing installation: six 1.17.0
Uninstalling six-1.17.0:
Successfully uninstalled six-1.17.0
Attempting uninstall: numpy
Found existing installation: numpy 2.0.2
Uninstalling numpy-2.0.2:
Successfully uninstalled numpy-2.0.2
Attempting uninstall: idna
Found existing installation: idna 3.11
Uninstalling idna-3.11:
Successfully uninstalled idna-3.11
Attempting uninstall: charset-normalizer
Found existing installation: charset-normalizer 3.4.4
Uninstalling charset-normalizer-3.4.4:
Successfully uninstalled charset-normalizer-3.4.4
Attempting uninstall: certifi
Found existing installation: certifi 2025.11.12
Uninstalling certifi-2025.11.12:
Successfully uninstalled certifi-2025.11.12
Attempting uninstall: scipy
Found existing installation: scipy 1.16.3
Uninstalling scipy-1.16.3:
Successfully uninstalled scipy-1.16.3
Attempting uninstall: requests
Found existing installation: requests 2.32.4
Uninstalling requests-2.32.4:
Successfully uninstalled requests-2.32.4
Attempting uninstall: pandas
Found existing installation: pandas 2.2.2
Uninstalling pandas-2.2.2:
Successfully uninstalled pandas-2.2.2
ERROR: pip's dependency resolver does not currently take into account all the packages that are installed. This behaviour is the source of the following dependency conflicts.
google-colab 1.0.0 requires pandas==2.2.2, but you have pandas 2.1.4 which is incompatible.
google-colab 1.0.0 requires requests==2.32.4, but you have requests 2.32.3 which is incompatible.
opencv-contrib-python 4.12.0.88 requires numpy<2.3.0,>=2; python_version >= "3.9", but you have numpy 1.26.4 which is incompatible.
google-adk 1.19.0 requires requests<3.0.0,>=2.32.4, but you have requests 2.32.3 which is incompatible.
shap 0.50.0 requires numpy>=2, but you have numpy 1.26.4 which is incompatible.
jaxlib 0.7.2 requires numpy>=2.0, but you have numpy 1.26.4 which is incompatible.
mizani 0.13.5 requires pandas>=2.2.0, but you have pandas 2.1.4 which is incompatible.
tsfresh 0.21.1 requires scipy>=1.14.0; python_version >= "3.10", but you have scipy 1.13.1 which is incompatible.
plotnine 0.14.5 requires pandas>=2.2.0, but you have pandas 2.1.4 which is incompatible.
pytensor 2.35.1 requires numpy>=2.0, but you have numpy 1.26.4 which is incompatible.
jax 0.7.2 requires numpy>=2.0, but you have numpy 1.26.4 which is incompatible.
opencv-python 4.12.0.88 requires numpy<2.3.0,>=2; python_version >= "3.9", but you have numpy 1.26.4 which is incompatible.
xarray 2025.11.0 requires pandas>=2.2, but you have pandas 2.1.4 which is incompatible.
opencv-python-headless 4.12.0.88 requires numpy<2.3.0,>=2; python_version >= "3.9", but you have numpy 1.26.4 which is incompatible.
Successfully installed certifi-2024.7.4 charset-normalizer-3.3.2 idna-3.7 numpy-1.26.4 pandas-2.1.4 pycatch22-0.4.5 pytz-2024.1 rarfile-4.2 requests-2.32.3 scipy-1.13.1 six-1.16.0 tzdata-2024.1 urllib3-2.2.2
1. Signal Processing: Unlocking Insights from Vibration Data¶
Vibration data from rotating machinery is a goldmine of information, but it's hidden in noise and complexity. Signal processing is the key to unlocking this information. Think of it as translating the machine's "language" of vibrations into a clear, understandable message about its health.
We start with the signal itself—a representation of vibration over time, either as a continuous analog wave or a discrete digital sequence. Signal processing then uses various techniques to enhance and analyze this signal:
- Time-domain analysis: Looking directly at the signal's shape over time. Think of it like reading a story—we look for sudden changes, repeating patterns, or unusual spikes.
- Frequency-domain analysis (FFT): Transforming the signal to see its hidden frequencies. This is like musical analysis—we identify the dominant notes (frequencies) that reveal the machine's internal workings and potential problems.
- Filtering: Cleaning up the signal by removing unwanted noise. This is like editing a recording—we remove background noise to hear the main instrument clearly.
By combining these techniques, we can extract crucial features from the vibration data, such as characteristic frequencies associated with specific faults, enabling early detection and diagnosis of problems. This allows for proactive maintenance, preventing costly breakdowns and ensuring operational efficiency. This notebook explores these techniques and how they are applied using [Package Name] to analyze vibration data and improve rotating machinery reliability.
2. Essential Theory¶
2.1 Sampling of Vibration Signals¶
Real-world vibration signals are continuous-time and analog. Digital processing requires sampling—converting the signal into a discrete-time sequence by measuring amplitude at regular intervals determined by the sampling frequency, fs, with sampling interval Ts = 1/fs.
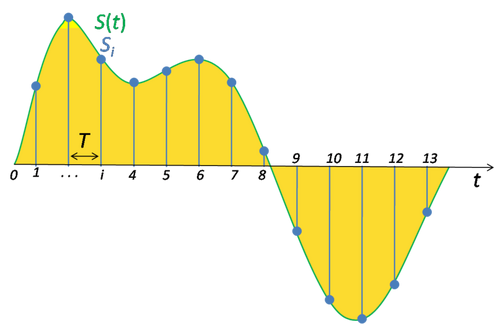
2.2 Nyquist-Shannon Sampling Theorem¶
Aliasing is phenomena which causes higher frequencies to appear as lower ones in the sampled data.

To avoid information loss (aliasing), the sampling frequency must be at least twice the maximum frequency present in the signal (fs ≥ 2fmax). Anti-aliasing filters are used before sampling to remove frequencies above fmax.
2.3 Leakage Error¶
Leakage error arises in frequency spectrum analysis when the analyzed signal's duration is not an integer multiple of its fundamental period. This discontinuity at the observation window's edges causes spectral energy to "leak" into adjacent frequency bins, blurring the true frequency components and distorting their amplitudes.
Windowing techniques (e.g., Hanning, Hamming) mitigate this by smoothly tapering the signal's ends, reducing the discontinuity and minimizing spectral leakage.
2.4 Modulation and Demodulation¶
Modulation modifies a carrier signal's properties using a message signal. In Amplitude Modulation (AM), the modulated signal is: s(t) = Ac[1 + m(t)]cos(2πfct), where Ac and fc are the carrier's amplitude and frequency, and m(t) is the message signal.
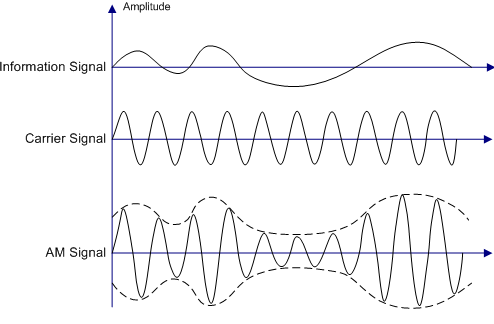
In RCM, machine speed variations modulate fault frequencies. Demodulation thecniques (etc. envelope extraction using Hilbert transform) recovers the message signal.
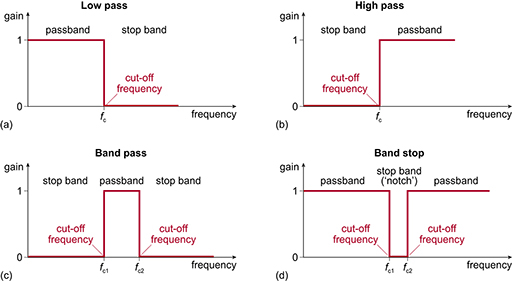
2.5 Frequency Filtering¶
Filtering selectively removes or attenuates specific frequency components. Common filter types include:
- Low-pass filters: Pass frequencies below a cutoff frequency and attenuate higher frequencies.
- High-pass filters: Pass frequencies above a cutoff frequency and attenuate lower frequencies.
- Band-pass filters: Pass frequencies within a specific band and attenuate frequencies outside this band.
- Band-stop filters (notch filters): Attenuate frequencies within a specific band and pass frequencies outside this band.
3. Signal Processing using Damavand¶
Damavand offers two main approaches to process vibration signals:
- Applying transformations: Damavand facilitates the application of the most frequent signal processing transformations.
- Fetaure extraction: In addition to the transformations, Damavand also supports the extraction of expert-defined features.
3.1 Transformations¶
3.1.0 Downloading and Mining the MFPT dataset¶
from damavand.damavand.datasets.downloaders import read_addresses, ZipDatasetDownloader
from damavand.damavand.datasets.digestors import MFPT
import pandas as pd
# Downloading the MFPT dataset
addresses = read_addresses()
downloader = ZipDatasetDownloader(addresses['MFPT'])
downloader.download_extract('MFPT.zip', 'MFPT/')
mfpt = MFPT('MFPT/', [
'baseline_1.mat',
'InnerRaceFault_vload_1.mat',
'InnerRaceFault_vload_2.mat',
'InnerRaceFault_vload_4.mat',
'InnerRaceFault_vload_7.mat',
'OuterRaceFault_1.mat',
'OuterRaceFault_vload_1.mat',
'OuterRaceFault_vload_2.mat',
'OuterRaceFault_vload_4.mat',
'OuterRaceFault_vload_7.mat',
])
# Mining the dataset
mining_params = {
97656: {'win_len': 16671, 'hop_len': 2000},
48828: {'win_len': 8337, 'hop_len': 1000},
}
mfpt.mine(mining_params)
# Signal/Metadata split
df = pd.concat(mfpt.data[48828]).reset_index(drop = True)
signals, metadata = df.iloc[:, : - 4], df.iloc[:, - 4 :]
3.1.1 Envelope extraction (Hilbert transform)¶
Extracting the envelope of of signals.
from damavand.damavand.signal_processing.transformations import env
# Envelope extraction
signals_env = env(signals)
3.1.2 Frequency spectrum extraction (Fast-Fourier Transform)¶
Applying the Fast-Fourier Transform algorithim to derive frequency domain representation of a set of signals
from scipy.signal.windows import hann
from scipy.signal import butter
from damavand.damavand.signal_processing.transformations import fft
# Defining a window to avoid frequency leakage
window = hann(signals_env.shape[1])
# Defining a bandpass frequency filter to both remove near-DC component and avoid aliasing
freq_filter = butter(25, [5, 23500], 'bandpass', fs = 48828, output='sos')
# Frequency spectra extraction, through FFT
signals_fft = fft(signals, freq_filter = freq_filter, window = window)
3.1.3 Refined frequency range spectrum extraction (ZoomFFT Algorithm)¶
Applying the ZoomFFT algorithm to derive a fine-grained frequency representation in a desired frequency range
from scipy.signal.windows import hann
from scipy.signal import butter
from damavand.damavand.signal_processing.transformations import zoomed_fft
# Defining a window to avoid frequency leakage
window = hann(signals_env.shape[1])
# Defining a bandpass frequency filter to both remove near-DC component and avoid aliasing
freq_filter = butter(25, [5, 23500], 'bandpass', fs = 48828, output='sos')
# Frequency spectra extraction within the range of 0 to 2500 Hz, through zoomed_FFT
signals_ZoomedFFT = zoomed_fft(signals_env, 0, 2500, 2500, 48828, freq_filter = freq_filter, window = window)
3.1.4 Time-Frequency representation extraction (Short-Time Fourier Transform)¶
Application of Short-Time Fourier Transform to derive Time-Frequency representation of the inputted signals
from scipy.signal.windows import hann
from scipy.signal import butter
from damavand.damavand.signal_processing.transformations import stft
# Defining a window to avoid frequency leakage (unlike previous transformations, the lenght of the window must match the window_len of the stft)
STFT_window = hann(2400)
# Defining a bandpass frequency filter to both remove near-DC component and avoid aliasing
STFT_freq_filter = butter(25, [5, 23500], 'bandpass', fs = float(metadata.iloc[0, 0]), output='sos')
# Time-Frequency representation extraction using 2400-point long segments and a hop lenght of 200 points.
signals_STFT = stft(signals, 2400, 200, STFT_freq_filter, STFT_window)
The following cells visualize the same observation, under different transformations.
import seaborn as sns
from matplotlib import pyplot as plt
sns.set()
from damavand.damavand.utils import *
fig, axes = plt.subplots(4, 1, figsize = (16, 10))
sns.lineplot(ax=axes[0], x=range(len(signals.iloc[0,:])), y = signals.iloc[0,:])
axes[0].set_title("Original Time Signal")
axes[0].set_ylabel("Amplitude")
axes[0].set_xlabel("sample")
axes[0].set_xlim(0, 8337)
sns.lineplot(ax=axes[1], x=range(len(signals_env.iloc[0,:])), y = signals_env.iloc[0, :])
axes[1].set_title("Envelope")
axes[1].set_ylabel("Amplitude")
axes[1].set_xlabel("sample")
axes[1].set_xlim(0, 8337)
sns.lineplot(ax=axes[2], x = fft_freq_axis(8337, 48828), y = signals_fft.iloc[0, :])
axes[2].set_title("FFT")
axes[2].set_ylabel("Amplitude")
axes[2].set_xlabel("Frequency (Hz)")
axes[2].set_xlim(0, 24424)
sns.lineplot(ax=axes[3], x = zoomed_fft_freq_axis(0, 2500, 2500), y = signals_ZoomedFFT.iloc[0, :])
axes[3].set_title("Zoomed FFT")
axes[3].set_ylabel("Amplitude")
axes[3].set_xlabel("Frequency (Hz)")
axes[3].set_xlim(0, 2500)
plt.subplots_adjust(hspace = 0.75)
fig.show()
import numpy as np
from damavand.damavand.utils import fft_freq_axis
t = np.linspace(0, 0.1707, 30)
f = fft_freq_axis(2400, 48828)
fig, ax = plt.subplots(figsize = (16, 8))
ax = sns.heatmap(signals_STFT[0, :, :], xticklabels = np.round(f, decimals = 2), yticklabels = np.round(t, decimals = 2), annot = False, cbar = False)
ax.set(xlabel = 'Frequency (Hz)', ylabel = 'Time (sec)')
ax.set_title('STFT')
ax.set_xticks(ax.get_xticks()[::30])
ax.set_yticks(ax.get_yticks()[::2])
fig.show()
Hand-crafted features (from both time and frequency domains) are widely used for rotating machinery conidition monitoring. Damavand, facilitates the extraction of such features from raw (time and frequency) data.
Features must be implemented as a Python function; then, they must be passed as key-vlaue pairs of "feature_name": (feature_function, (args), (kwargs)) to the feature_extractor function, alongside the signal bank.
The following section, demonstrate the extraction of both time and frequency domains, respectively.
3.2.1 Time-domain Features¶
from damavand.damavand.signal_processing.feature_extraction import *
from numpy import mean, std
from scipy.stats import skew, kurtosis
# Defining the desired features (below is a wide set of time-domain features)
time_features = {
'mean': (mean, (), {}),
'std': (std, (), {}),
'smsa': (smsa, (), {}),
'rms': (rms, (), {}),
'peak': (peak, (), {}),
'skew': (skew, (), {}),
'kurtosis': (kurtosis, (), {}),
'crest_factor': (crest_factor, (), {}),
'clearance_factor': (clearance_factor, (), {}),
'shape_factor': (shape_factor, (), {}),
'impulse_factor': (impulse_factor, (), {}),
}
# Extracting the features from the signals
time_features_df = feature_extractor(signals, time_features)
time_features_df
| mean | std | smsa | rms | peak | skew | kurtosis | crest_factor | clearance_factor | shape_factor | impulse_factor | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | -0.233352 | 1.843121 | 0.699720 | 1.857834 | 30.15747 | -0.550328 | 39.490115 | 16.232595 | 43.099347 | 1.891972 | 30.711608 |
| 1 | -0.234489 | 1.720509 | 0.692389 | 1.736415 | 30.15747 | -0.538847 | 36.334443 | 17.367663 | 43.555657 | 1.814836 | 31.519462 |
| 2 | -0.234101 | 1.901948 | 0.708425 | 1.916301 | 32.18819 | -0.751920 | 47.621448 | 16.797046 | 45.436280 | 1.924807 | 32.331076 |
| 3 | -0.235983 | 1.797137 | 0.686839 | 1.812564 | 32.18819 | -0.626101 | 44.018940 | 17.758374 | 46.864249 | 1.891778 | 33.594892 |
| 4 | -0.234448 | 1.800635 | 0.690554 | 1.815833 | 32.18819 | -0.547199 | 43.198745 | 17.726401 | 46.612102 | 1.883161 | 33.381664 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 1107 | -0.156233 | 1.504572 | 0.716416 | 1.512662 | 16.14121 | 0.375640 | 14.036173 | 10.670731 | 22.530485 | 1.610511 | 17.185332 |
| 1108 | -0.157083 | 1.534541 | 0.723250 | 1.542560 | 16.14121 | 0.404746 | 14.021135 | 10.463912 | 22.317611 | 1.621624 | 16.968533 |
| 1109 | -0.157834 | 1.468633 | 0.713651 | 1.477090 | 16.14121 | 0.327941 | 13.254681 | 10.927712 | 22.617794 | 1.586590 | 17.337794 |
| 1110 | -0.159280 | 1.455467 | 0.711277 | 1.464156 | 16.14121 | 0.284900 | 13.136450 | 11.024239 | 22.693293 | 1.581841 | 17.438593 |
| 1111 | -0.160429 | 1.499163 | 0.724039 | 1.507723 | 16.14121 | 0.253219 | 13.184033 | 10.705689 | 22.293274 | 1.594859 | 17.074060 |
1112 rows × 11 columns
3.2.2 Frequency-domain Features¶
from scipy.signal.windows import hann
from scipy.signal import butter
from numpy import mean, var
from scipy.stats import skew, kurtosis
from damavand.damavand.signal_processing.feature_extraction import *
from damavand.damavand.utils import *
# Applying the FFT to transform data into frequency-domain
window = hann(signals.shape[1])
freq_filter = butter(25, [5, 12500], 'bandpass', fs = 25600, output='sos')
signals_fft = fft(signals, freq_filter = freq_filter, window = window)
# Extracting frequency axis, as it is essential for some of the features
freq_axis = fft_freq_axis(8337, 48828)
# Defining the desired features (below is a wide set of frequency-domain features)
freq_features = {
'mean': (mean, (), {}),
'var': (var, (), {}),
'skew': (skew, (), {}),
'kurtosis': (kurtosis, (), {}),
'spectral_centroid': (spectral_centroid, (freq_axis,), {}),
'P17': (P17, (freq_axis,), {}),
'P18': (P18, (freq_axis,), {}),
'P19': (P19, (freq_axis,), {}),
'P20': (P20, (freq_axis,), {}),
'P21': (P21, (freq_axis,), {}),
'P22': (P22, (freq_axis,), {}),
'P23': (P23, (freq_axis,), {}),
'P24': (P24, (freq_axis,), {}),
}
# Extracting the features from the frequency domain signals (assuming that frequency domain signals are stored in signals_fft)
freq_features_df = feature_extractor(signals_fft, freq_features)
freq_features_df
/content/damavand/damavand/signal_processing/feature_extraction.py:103: RuntimeWarning: invalid value encountered in sqrt return np.mean(np.sqrt(np.subtract(freq_axis, spectral_centroid(spectrum, freq_axis))) * spectrum) / np.sqrt(P17(spectrum, freq_axis))
| mean | var | skew | kurtosis | spectral_centroid | P17 | P18 | P19 | P20 | P21 | P22 | P23 | P24 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.018065 | 0.000212 | 1.426027 | 2.114200 | 11053.444596 | 796.261629 | 12540.952280 | 2.935690e+08 | 0.731940 | 0.072037 | 2.205723 | 125.363550 | 0.035847 |
| 1 | 0.016538 | 0.000166 | 1.440692 | 2.510849 | 11015.384364 | 790.490364 | 12614.367865 | 3.038027e+08 | 0.723718 | 0.071762 | 2.306647 | 130.261142 | 0.033860 |
| 2 | 0.015911 | 0.000163 | 1.544936 | 2.872723 | 10796.428140 | 779.206239 | 12438.754858 | 3.016914e+08 | 0.716136 | 0.072173 | 2.514890 | 136.998907 | 0.032348 |
| 3 | 0.016080 | 0.000178 | 1.584992 | 2.905862 | 10532.076594 | 767.281209 | 12146.457376 | 2.907104e+08 | 0.712393 | 0.072852 | 2.662891 | 142.330173 | 0.031666 |
| 4 | 0.017501 | 0.000236 | 1.647699 | 2.950443 | 10594.040230 | 773.264096 | 12099.593080 | 2.786391e+08 | 0.724853 | 0.072990 | 2.140161 | 136.694506 | 0.034301 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 1107 | 0.015128 | 0.000206 | 2.559033 | 10.592382 | 9658.153861 | 723.161190 | 11307.004650 | 2.685357e+08 | 0.689996 | 0.074876 | 3.546391 | 158.120703 | 0.029296 |
| 1108 | 0.014921 | 0.000198 | 2.563811 | 11.058002 | 9697.088412 | 723.418752 | 11362.562278 | 2.705444e+08 | 0.690807 | 0.074602 | 3.459591 | 157.621090 | 0.029264 |
| 1109 | 0.014107 | 0.000178 | 2.703421 | 13.286557 | 9592.960582 | 712.859411 | 11315.835834 | 2.738293e+08 | 0.683828 | 0.074311 | 3.729962 | 165.473946 | 0.027922 |
| 1110 | 0.013595 | 0.000168 | 2.972876 | 16.970993 | 9487.339448 | 700.212027 | 11228.315022 | 2.738700e+08 | 0.678488 | 0.073805 | 4.045796 | 173.713123 | 0.026819 |
| 1111 | 0.013689 | 0.000173 | 3.137925 | 19.340805 | 9503.659572 | 696.582752 | 11214.560026 | 2.722860e+08 | 0.679625 | 0.073296 | 4.115159 | 175.286662 | 0.026755 |
1112 rows × 13 columns
3.2.3 catch-22 Feature Set¶
Authors of this study introduce a feature set that not only provide strong classification performance over a wide range of time series dataset, but also are minimally redundant. Using the official implementation of the authors under the hood, the catch22_features function extract this feature set.
from damavand.damavand.datasets.downloaders import read_addresses, ZipDatasetDownloader
from damavand.damavand.datasets.digestors import MFPT
from damavand.damavand.signal_processing.feature_extraction import *
from damavand.damavand.signal_processing.transformations import fft
from damavand.damavand.utils import *
from zipfile import ZipFile
import pandas as pd
# Downloading the MFPT dataset
addresses = read_addresses()
downloader = ZipDatasetDownloader(addresses['MFPT'])
downloader.download_extract('MFPT.zip', 'MFPT/')
mfpt = MFPT('MFPT/', [
'baseline_1.mat',
'InnerRaceFault_vload_1.mat',
'InnerRaceFault_vload_2.mat',
'InnerRaceFault_vload_4.mat',
'InnerRaceFault_vload_7.mat',
'OuterRaceFault_1.mat',
'OuterRaceFault_vload_1.mat',
'OuterRaceFault_vload_2.mat',
'OuterRaceFault_vload_4.mat',
'OuterRaceFault_vload_7.mat',
])
# Mining the dataset
mining_params = {
97656: {'win_len': 16671, 'hop_len': 2000},
48828: {'win_len': 8337, 'hop_len': 1000},
}
mfpt.mine(mining_params)
# Signal/Metadata split
df = pd.concat(mfpt.data[48828]).reset_index(drop = True)
signals, metadata = df.iloc[:, : - 4], df.iloc[:, - 4 :]
# Toggle the include_additionals to False, to stay with the original 22 features
catch22_features_df = catch22_features(signals, include_additionals=True)
catch22_features_df
| DN_HistogramMode_5 | DN_HistogramMode_10 | CO_f1ecac | CO_FirstMin_ac | CO_HistogramAMI_even_2_5 | CO_trev_1_num | MD_hrv_classic_pnn40 | SB_BinaryStats_mean_longstretch1 | SB_TransitionMatrix_3ac_sumdiagcov | PD_PeriodicityWang_th0_01 | ... | DN_OutlierInclude_n_001_mdrmd | SP_Summaries_welch_rect_area_5_1 | SB_BinaryStats_diff_longstretch0 | SB_MotifThree_quantile_hh | SC_FluctAnal_2_rsrangefit_50_1_logi_prop_r1 | SC_FluctAnal_2_dfa_50_1_2_logi_prop_r1 | SP_Summaries_welch_rect_centroid | FC_LocalSimple_mean3_stderr | DN_Mean | DN_Spread_Std | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | -1.132135 | 0.378111 | 0.818324 | 2 | 0.021987 | 0.713018 | 0.931262 | 16.0 | 0.000533 | 3 | ... | -0.051337 | 0.155494 | 7.0 | 2.165575 | 0.62 | 0.24 | 1.254029 | 1.280226 | -0.233352 | 1.843231 |
| 1 | -1.212156 | 0.405717 | 0.838842 | 2 | 0.015240 | 0.417357 | 0.935940 | 17.0 | 0.000571 | 4 | ... | -0.093559 | 0.184471 | 8.0 | 2.165307 | 0.30 | 0.88 | 1.243675 | 1.265813 | -0.234489 | 1.720612 |
| 2 | -1.630546 | -0.113629 | 0.789737 | 2 | 0.021483 | 1.812335 | 0.929822 | 17.0 | 0.000723 | 3 | ... | -0.027228 | 0.149193 | 8.0 | 2.167602 | 0.34 | 0.88 | 1.273971 | 1.274452 | -0.234101 | 1.902062 |
| 3 | -2.033294 | -0.458779 | 0.778984 | 2 | 0.016595 | 1.561510 | 0.930422 | 17.0 | 0.000541 | 3 | ... | 0.009116 | 0.167291 | 8.0 | 2.168096 | 0.32 | 0.82 | 1.315772 | 1.257946 | -0.235983 | 1.797245 |
| 4 | -2.030197 | -0.458741 | 0.780336 | 2 | 0.015508 | 1.673901 | 0.929343 | 17.0 | 0.000431 | 3 | ... | -0.011875 | 0.177576 | 8.0 | 2.164446 | 0.26 | 0.68 | 1.318840 | 1.251895 | -0.234448 | 1.800743 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 1107 | -0.677851 | 0.316727 | 1.157365 | 2 | 0.037245 | 0.167977 | 0.947937 | 19.0 | 0.001133 | 3 | ... | -0.024829 | 0.365049 | 10.0 | 2.114870 | 0.20 | 0.88 | 0.918471 | 1.153364 | -0.156233 | 1.504663 |
| 1108 | -0.664059 | 0.311096 | 1.126916 | 2 | 0.037907 | 0.277817 | 0.947817 | 19.0 | 0.001116 | 3 | ... | -0.109872 | 0.343456 | 10.0 | 2.115158 | 0.24 | 0.88 | 0.927291 | 1.165169 | -0.157083 | 1.534633 |
| 1109 | -0.693349 | 0.325568 | 1.198491 | 2 | 0.030415 | 0.134321 | 0.947817 | 19.0 | 0.001211 | 3 | ... | -0.053257 | 0.394248 | 10.0 | 2.112970 | 0.20 | 0.12 | 0.906199 | 1.136620 | -0.157834 | 1.468721 |
| 1110 | -0.698628 | 0.329506 | 1.190782 | 2 | 0.029601 | 0.083865 | 0.947457 | 14.0 | 0.000810 | 3 | ... | 0.011455 | 0.392137 | 10.0 | 2.119922 | 0.20 | 0.12 | 0.904665 | 1.134735 | -0.159280 | 1.455554 |
| 1111 | -0.677498 | 0.320668 | 1.171436 | 2 | 0.034213 | -0.004840 | 0.948297 | 14.0 | 0.000821 | 3 | ... | 0.031246 | 0.384824 | 10.0 | 2.120204 | 0.20 | 0.12 | 0.915403 | 1.144036 | -0.160429 | 1.499253 |
1112 rows × 24 columns